Thursday, September 3, 2020
Science and engineering researchers at UC Santa Barbara are already reaping the rewards of a new, state-of-the-art graphics processing unit (GPU) computer cluster that was installed on campus this month. Originally designed to advance video games and on-screen three-dimensional rendering, GPUs are now powering scientific data processing and academic research, dramatically accelerating modeling and computations. Because it is specifically designed for basic mathematical operations, for some codes, a GPU-based system can perform computations thirty to fifty times faster than conventional central processing unit (CPU) hardware can. GPUs are more efficient — performing multiple calculations simultaneously, while CPUs work through calculations a few at a time — and as a result have turbocharged computational research in a growing number of fields ranging from materials characterization and molecular simulation, to astrophysics and machine learning.
“Many GPUs are required to perform large-scaled computations, and this new cluster greatly expands the number of the highest-performing GPUs that are openly available to all members of the campus community, from 12 to 52,” said Kris Delaney, a co-principal investigator on the grant and project scientist with the Materials Research Laboratory (MRL). “This is a game-changer for researchers at UCSB.”
The Center for Scientific Computing (CSC) at the California NanoSystems Institute (CNSI) and the MRL is managing and maintaining the new system, which was funded by a $400,000 grant from the National Science Foundation to accelerate scientific computing workloads. The upgrade includes twelve compute nodes, with each one supporting four high-end NVIDIA Tesla V100 GPUs that are equipped with hardware double-precision processing, large (32 GB) memory, and NVLink for fast inter-GPU communications. Ten of the nodes were added to the existing High Performance Computing (HPC) campus cluster “Pod” that has three GPU nodes as part of its complement of computing nodes. The HPC cluster is currently available to UCSB researchers. The two remaining nodes became part of the delocalized Pacific Research Platform Nautilus cluster, headquartered at UC San Diego, which connects a few dozen universities along the West Coast including UCSB. CSC staff plan to continue hosting seminars, workshops, and webinars to help attain proficiency with the new cluster.
“The improvements of GPUs are happening very fast, so we need to be constantly upgrading equipment if we want to do the latest GPU-enabled research,” said Paul Weakliem, CSC director. “Five years ago, there was a fairly small set of people who could use GPUs. Since then, the number of research fields using them has increased dramatically, and as a result, the demand has been rising quickly, too.”

Fuzzy Rogers, CSC co-director, adds, “With many researchers restricted from working in their labs due to the COVID-19 pandemic, there has also been an increase in demand as they explore their research with new computational techniques.”
Faster computational speeds not only enable higher throughput, but also opportunities to address new research questions. For example, the expansion will enhance efforts to develop new bio-based materials in the new BioPolymers, Automated Cellular Infrastructure, Flow, and Integrated Chemistry: Materials Innovation Platform (BioPACIFIC MIP), a collaboration between UCSB and UC Los Angeles established in August with a $23.7 million NSF grant.
“These computing resources add to a critical mass of infrastructure and expertise that enable BioPACIFIC stakeholders to leverage the CSC in numerous strategic and scientific endeavors,” said Tal Margalith, CNSI’s executive director of technology and executive director of UCSB’s BioPACIFIC MIP operation. “The cluster and CSC will support the computation, simulation, and data science aspects of BioPACIFIC, and they will be critical to the project’s success.”
The new cluster will also drive the development and application of novel computational research techniques in three specific projects associated with the grant.
One project, led by Delaney, is intended to develop a unique simulation methodology to predict the structures and properties of complex polymers formulations found in various consumer products, including cosmetics, liquid soaps, detergents, paints, food, and motor oil. Such products are made from a complicated list of ingredients, which self-assemble to determine their properties and functions. The methodology, which also supports efforts of the BioPACIFIC MIP, bridges pioneering simulation techniques developed by two UCSB chemical engineering research groups: molecular and coarse-grained modeling techniques developed by Professor M. Scott Shell, and polymer field theory used by Professor Glenn Fredrickson to simulate at the meso-scale. Simulations will be used to model the molecular structures and chemistry of new materials and suggest how design criteria can be met.
“Even with our novel approach to multi-scale modeling, the simulations required to meet our goal of predicting structures and properties from chemical formulation are large and computationally intensive,” said Delaney. “By using cutting-edge GPUs as the computing hardware, very large simulations become more manageable.”
The new computing resources have also been directly coupled to a revolutionary microscope designed by materials professor Tresa Pollock and now being commercially manufactured by Thermo Fisher Scientific, the world’s leading microscope manufacturer.
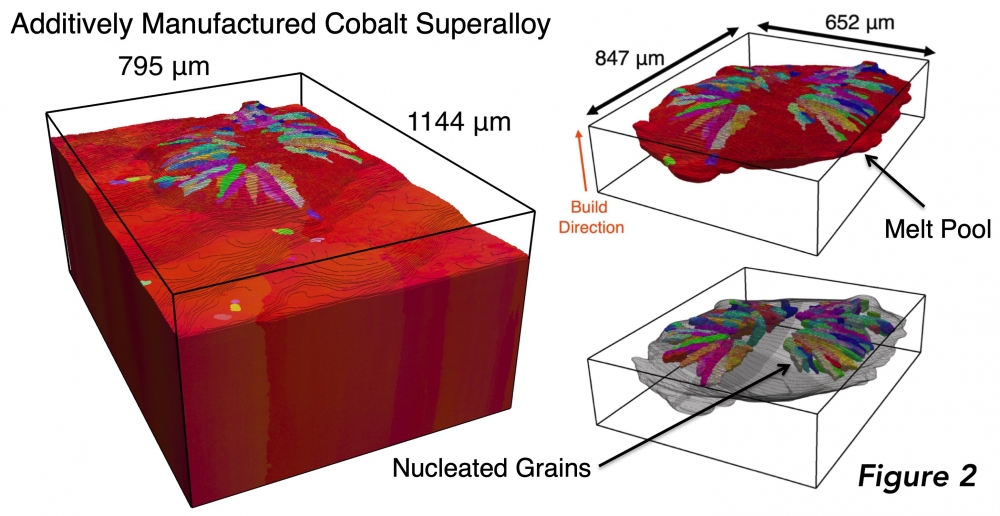
Pollock’s research group focuses on the discovery, early development, and improvement of structural materials designed for extreme environments, such as cobalt superalloys that are used in aerospace applications. The group developed the TriBeam microscope to gather information about the microstructure and defects that exist in materials by using a femtosecond laser to remove layers of materials at rates three to five orders of magnitude faster than existing instruments. Examining microstructures is important because they control a material’s properties that factor into its performance in extreme conditions. The new computer cluster will help process the terabytes of data generated by the TriBeam for each dataset.
“We will be able to perform on-the-fly processing and reconstruction of the 3D data generated,” said McLean Echlin, a research scientist in Pollock’s group. “These new processing capabilities will increase the fidelity of the data gathered and provide feedback for real-time instrument control, which will allow us to hone in on specific property-controlling features in materials.”
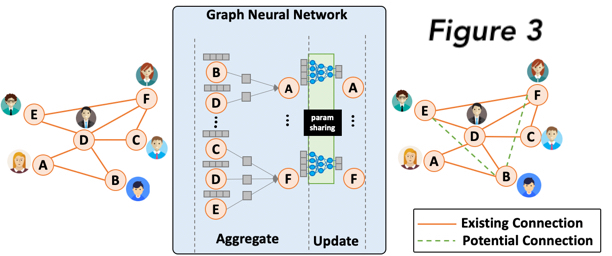
The cluster has also allowed Yufei Ding, an assistant professor in the Computer Science Department, to process data that would normally not be possible in her research on machine learning and quantum computing. Ding’s group aims to facilitate faster and more energy-efficient neural-network training and inference by minimizing the size of the network — referred to as “dynamic pruning” — and by modeling the associated architecture and framework. A neural network is a series of algorithms that recognizes relationships in a set of data through a process that resembles the way the human brain operates. Training a neural network is challenging because it involves using an optimization algorithm to find the best weight, or strength, of a connection between inputs and outputs. Ding’s work to speed up neural network training could impact the way machine learning is applied to autonomous driving, computer vision, and speech recognition. Her group also studies quantum computing, investigating advanced simulation schemes and debugging tools, in order to make quantum programming easier to perform.
“This new GPU cluster enables a tremendous amount of research to be completed that would not be done or would just take too much time to fulfill,” said Ding, whose research has resulted in papers that were already accepted in five top conference publications. “My work on neural architecture search for machine learning and simulator constructor for quantum computing require GPU systems that have both massive computation units and a large amount of on-board memory.”
The ripple effect of expanding the GPUs available to researchers on campus will reach far beyond faculty. The expanded Pod HPC cluster will also power the training of the next generation of scientists and engineers. Based on current usage, the center estimates that up to three-fourths of the users of the expanded resources will be undergraduate and graduate students. The shared resource will also be available to high school and community college students and teachers who participate in campus-sponsored programs such as the MRL's Research Experience for Teachers (RET) program and the California Alliance for Minority Participation (CAMP) program, as well as the Center for Science and Engineering Partnerships’ (CSEP’s) Maximizing Access to Research Careers (MARC) and Summer Institute in Mathematics and Sciences (SIMS).
Captions
Figure 1: Schematic of the hierarchy of simulation approaches that are combined to predict mesoscale structure and properties of complex polymeric solution formulations. Beginning with atomistic molecular dynamics (left), small-scale simulations are used with relative-entropy coarse-graining to parameterize models that can be converted via an analytic, exact transformation to a field-theory, which is then simulated with field-theoretic approaches (e.g., complex Langevin sampling) to predict meso-scale structures and properties. Both the particle and field simulation methods are able to take advantage of GPU hardware to dramatically increase the size and throughput of simulations.
Figure 2: A TriBeam 3D dataset of an additively manufactured cobalt-base superalloy showing a single melt pool on the dataset surface and the grain structure that nucleates within it during the 3D printing process. Datasets such as these are reprocessed for higher data fidelity using the new compute resources on the pod cluster.
Figure 3: Graph Neural Networks (GNNs) aims at exploiting the potential connection among objects by extending machine learning to graph data. Based on a relation graph (left) among users, we can apply GNN (middle) on this user-relation graph to generate a feature vector (embedding) for each user that can capture the user specialty. After getting the users’ embeddings, we can explore the potential relations among users based on their embedding similarity, also known as link prediction. GNNs are able to take advantage of massively parallelized GPUs to dramatically improve their performance. Especially, by levering GPU-pod clusters for GNN training, we reduce the original GNN training time by 50%.
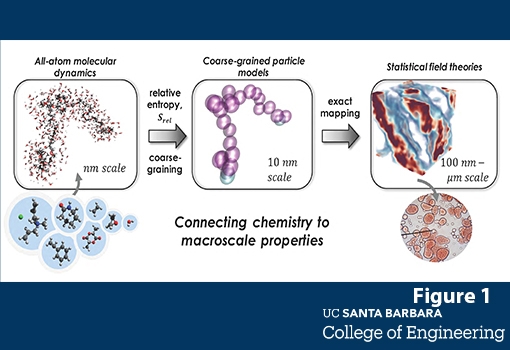
Schematic of the hierarchy of simulation approaches that are combined to predict mesoscale structure and properties of complex polymeric solution formulations.
